The Role of Artificial Intelligence in Antibody Drug Discovery
Traditional antibody discovery is notoriously slow and resource-intensive, but a new wave of AI tools is enabling optimization of candidates prior to them reaching the clinic, accelerating development timelines.
By Mandar Bhonde, Appalaraju Jaggupilli, and Rajeshwari B R, all from Enzene.
Antibody-based therapeutics have revolutionized modern medicine through their remarkable specificity and efficacy in targeting cancer, autoimmune, and infectious diseases. More than 100 monoclonal antibodies (mAbs) have gained global clinical approval, reflecting their immense therapeutic demand (1,2). However, conventional discovery workflows, such as hybridoma generation, phage display, and animal immunization, remain slow, resource-intensive, and have high attrition (3,4). These limitations have catalyzed the adoption of artificial intelligence (AI) to reshape the antibody discovery paradigm (5,6).
From Experimental to Computational Discovery
Early experimental platforms, such as hybridoma and phage display, established the foundation for antibody generation but faced inherent constraints in throughput and humanization (3,4). The integration of computational biology introduced in silico tools, including RosettaAntibody, SnugDock, and RosettaAntibodyDesign (RAbD), and enabled structure modeling and affinity optimization (7–9). However, the true transformation within the space came with machine learning (ML) and deep learning (DL) architectures capable of learning directly from biological sequences and structures. Landmark advances, such as AlphaFold2 and RoseTTAFold, achieved near-experimental accuracy in protein structure prediction, dramatically reducing reliance on crystallography and enabling accurate modeling of antibody–antigen complexes (10–12).
Fast-Paced Discovery and Design
AI-driven discovery platforms now accelerate candidate generation by integrating sequence prediction, structure modeling, and binding affinity estimation (12,13). Transformer-based sequence models, such as AntiBERTa and AbLang, learn contextual dependencies across millions of antibody sequences, generating human-like variants with optimized complementarity-determining regions (CDRs) (14,15). Generative AI approaches using diffusion models and variational autoencoders enable de novo antibody design, reducing discovery timelines from years to weeks (16–18). Diffusion-based frameworks, such as RFdiffusion, have achieved atomic-level precision in designing highly specific antibodies, validated by cryo-electron microscopy (19).
Structural and Functional Optimization
AI tools support optimization of key developability attributes — stability, viscosity, aggregation risk, and immunogenicity (19,20). Platforms — examples of which include Therapeutic Antibody Profiler (TAP), and SOLart — incorporate multi-parameter models to predict manufacturability and biophysical performance early in discovery (21,22). This proactive assessment reduces late-stage failures and improves production feasibility. Graph-based models enhance paratope–epitope mapping, improving prediction of antibody–antigen interactions with sub-angstrom precision (6–8).
Integration with High-Throughput Screening
Coupling AI with next-generation sequencing (NGS) and automated laboratory platforms further accelerates discovery (1,23). AI identifies high-affinity and diverse sequences from vast antibody repertoires that might otherwise be overlooked (18,23). ML algorithms using feature embedding have predicted enrichment of rare clones and improved binding specificity (24). This synergy between AI prediction and experimental validation creates a continuous feedback loop supporting rational antibody engineering (18).
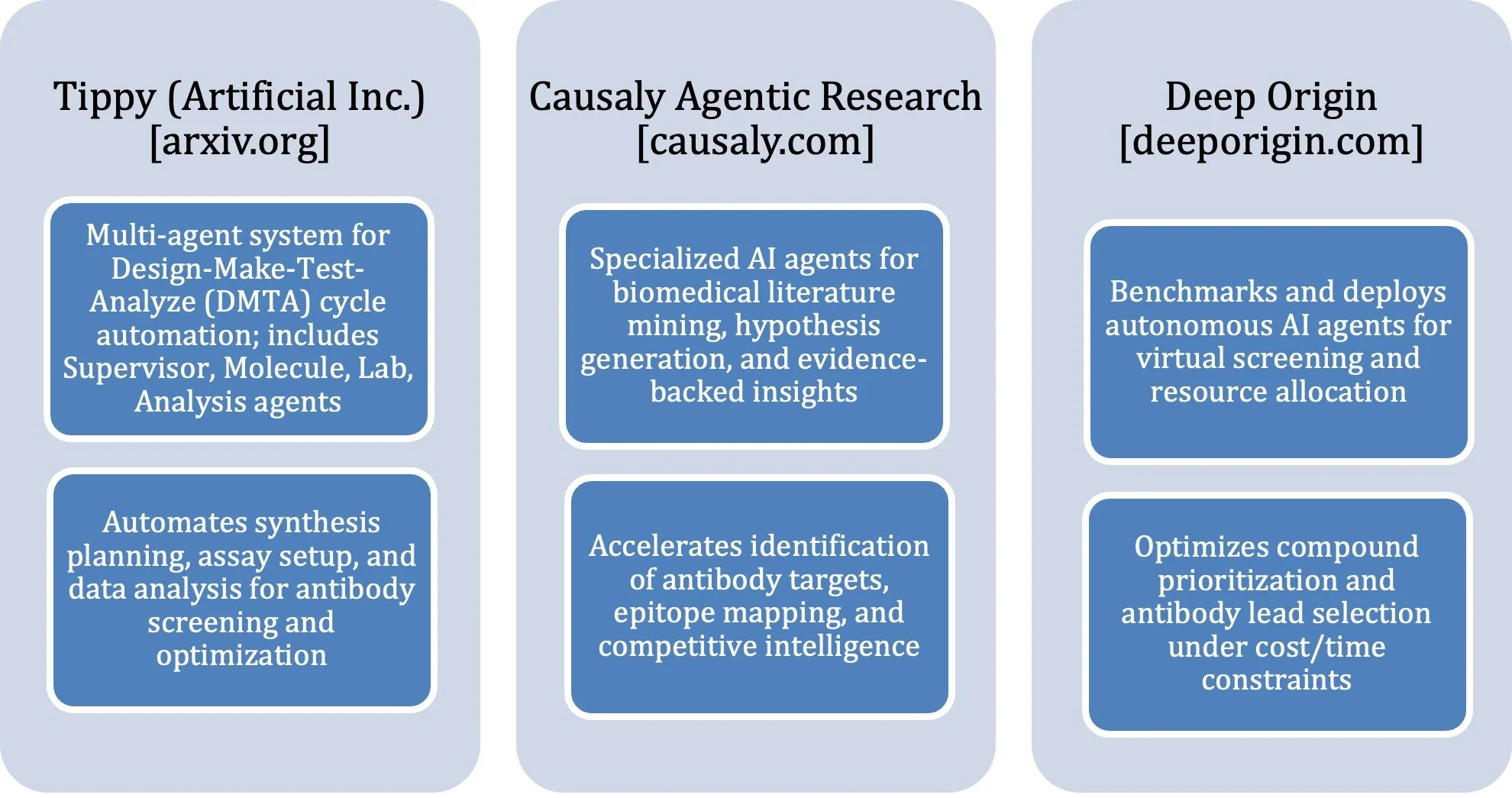
Figure 1 provides a snapshot of the Agentic AI tools and their applications in drug discovery.
Figure 1: Agentic AI tools for End-to-End Drug Discovery Solutions. [FIGURE IS COURTESY OF AUTHORS.]
Challenges and Future Prospects
Despite significant achievements, challenges persist. Public antibody databases lack paired heavy–light chain information and standardized assay metrics, limiting model generalization (6). Future progress depends on multimodal integration linking structural and omics data with physics-informed AI that merges molecular dynamics with neural networks. Emerging closed-loop discovery ecosystems, where AI designs and robotic systems test in real time, are ushering in autonomous biologic design (25).
The global antibody drug market, projected to surpass USD 445 billion within five years, will benefit enormously from these AI innovations. As Agentic AI systems combine literature mining and iterative optimization, discovery becomes faster and more democratized (Figure 1)(26). AI promises to expand antibody design beyond natural limitations, enabling development of bispecifics and antibody–drug conjugates for previously undruggable targets, heralding a new age of AI-orchestrated therapeutic discovery (6).
References
Lu, R.M.; Hwang, Y.C.; Liu, I.J.; et al. Development of Therapeutic Antibodies for the Treatment of Diseases. J. Biomed. Sci. 2020, 27, 1. DOI: 10.1186/s12929-019-0592-z.
Madej B.; Tomaszewski F.; Szmajda-Krygier D. et al. Monoclonal Antibodies: Historical Perspective and Current Trends in Biological Drug Development. Int. J. Mol. Sci. 2025, 26(18), 8794; DOI: 10.3390/ijms26188794.
Köhler, G.; Milstein, C. Continuous Cultures of Fused Cells Secreting Antibody of Predefined Specificity. Nature. 1975, 256, 495–497. DOI: 10.1038/256495a0.
Lu, R.M.; Chiang, H.; Yuan, J.P.; et al. Technological Advancements in Antibody-based Therapeutics for treatment of Diseases. J. Biomed. Sci. 2025, 32(1), 98. DOI: 10.1186/s12929-025-01190-2.
Mak, K.K.; Pichika, M.R. Artificial Intelligence in Drug Development: Present Status and Future Prospects. Drug Discov Today. 2019, 24(3), 773–780. DOI: 10.1016/j.drudis.2018.11.014.
Santuari, L.; Bachmann Salvy, M.; Xenarios, I.; Arpat, B. AI-Accelerated Therapeutic Antibody Development: Practical Insights. Front. Drug Discov. 2024, 4, 1447867. DOI:10.3389/fddsv.2024.1447867.
Lyskov, S.; Gray J.J. The RosettaDock Server for Local Protein–Protein Docking. Nucleic Acids Res. 2008, 70(3), 754–762. DOI: 10.1093/nar/gkn216.
Sircar, A.; Gray, J.J. SnugDock: Paratope Structural Optimization During Antibody–Antigen Docking Compensates for Errors in Antibody Homology Models. PLoS Comput. Biol. 2010, 6(1), e1000644. DOI: 10.1371/journal.pcbi.1000644.
Adolf-Bryfogle, J.; Kalyuzhniy, O.; Kubitz, M.; et al. RosettaAntibodyDesign (RAbD): A General Framework for Computational Antibody Design. PLoS Comput. Biol. 2018, 14 (4), e1006112. DOI: 10.1371/journal.pcbi.1006112.
Jumper, J.; Evans, R.; Pritzel, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature. 2021, 596(7873), 583–589. DOI: 10.1038/s41586-021-03819-2.
Baek, M.; Dimaio, F.; Anishchenko, I.; et al. Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science. 2021, 373(6557), 871–876. DOI: 10.1126/science.abj8754.
Zheng, J.; Wang Y.; Liang Q.; et al. The Application of Machine Learning on Antibody Discovery and Optimization. Molecules 2024, 29(24), 5923. DOI: 10.3390/molecules29245923.
Kavousipour, S.; Barazesh, M.; Mohammadi, S. Artificial Intelligence in Antibody Design and Development: Harnessing the Power of Computational Approaches. Med. Biol. Eng. Comput. 2025, 63, 3475-3501. DOI:10.1007/s11517-025-03429-4.
Choi, Y. Artificial Intelligence for Antibody Reading Comprehension: AntiBERTa. Patterns (N Y). 2022, 3(7), 100535. DOI: 10.1016/j.patter.2022.100535.
Olsen, T.H.; Moal, I.H.; Deane, C.M. AbLang: An Antibody Language Model for Completing Antibody Sequences. Bioinform Adv., 2022, 2(1), vbac046. DOI: 10.1093/bioadv/vbac046.
Chowdhury, R.; Bouatta, N.; Biswas, S.; et al. Single-Sequence Protein Structure Prediction Using a Language Model and Deep Learning. Nat. Biotechnol. 2022, 40(11), 1617-1623. DOI:10.1038/s41587-022-01432-w.
Liu, Y.; Zhang, L.; Jiang, Z. et al. Applications of Artificial Intelligence in Biotech Drug Discovery and Product Development. MedComm. 2020. 30, 6(8), e70317. DOI: 10.1002/mco2.70317.
Matsunaga, R.; Tsumoto, K. Accelerating Antibody Discovery and Optimization with High-Throughput Experimentation and Machine Learning. J. Biomed. Sci. 2025, 32(1), 46. DOI:10.1186/s12929-025-01141-x.
Bennett, N.R.; Watson, J.L.; Ragotte, R.J.; et al. Atomically Accurate De Novo Design of Antibodies with RFdiffusion. Nature. 2026, 649, 183–193. DOI:10.1038/s41586-025-09721-5.
Raybould, M.I.J.; Marks, C.; Krawczyk, K.; et al. Five Computational Developability Guidelines for Therapeutic Antibody Profiling. Proc Natl Acad Sci U S A. 2019, 116(10), 4025-4030. DOI: 10.1073/pnas.1810576116.
Hou, Q.; Kwasigroch, J.M.; Rooman, M.; et al. SOLart: A Structure-Based Method to Predict Protein Solubility and Aggregation. Bioinformatics. 2020, 36(5), 1445-1452. DOI: 10.1093/bioinformatics/btz773.
Khetan, R.; Curtis, R.; Deane, C.M.; et al. Current Advances in Biopharmaceutical Informatics: Guidelines, Impact and Challenges in the Computational Developability Assessment of Antibody Therapeutics. mAbs. 2022, 14(1). DOI: 10.1080/19420862.2021.2020082
Mason, D.M.; Friendensohn, S.; Weber, C.R.; et al. Deep Learning Enables Therapeutic Antibody Optimization in Mammalian Cells by Deciphering High-Dimensional Protein Sequence Space. bioRxiv. 2019, 617860.
Ruffolo, JA.; Sulam, J.; Gray, J.J. Antibody Structure Prediction Using Interpretable Deep Learning. Patterns (N Y). 2021, 3(2), 100406. DOI:10.1016/j.patter.2021.100406.
Karniadakis, G E.; Kevrekidis, I G.; Lu, L.; et al. Physics-Informed Machine Learning. Nat. Rev. Phy. 2021, 3, 422-440. DOI: 10.1038/s42254-021-00314-5.
Seal, S.; Huynh, D L.; Chelbi, M.; et al. AI Agents in Drug Discovery. arXiv. 2025, arXiv:2510.27130. DOI:10.48550/arXiv.2510.27130
About the Authors
Mandar Bhonde, Ph.D.
Sr. GM & Head, Discovery Ops
Dr. Mandar Bhonde is a drug discovery and development professional with more than 18 years of experience in small molecule R&D Operations. He has an established track record of leadership spanning functions such as discovery research to candidate characterization & out-licensing. Prior to joining Enzene, he held prominent roles at biopharmaceutical firms like Piramal Life Sciences Ltd., and Lupin Ltd., where he successfully led numerous programs across several therapeutic areas from discovery to IND.
Dr. Bhonde holds masters in Molecular Biology from Savitribai Phule Pune University (formerly University of Pune) and a PhD in Cancer Biology from Charite Campus Benjamin Franklin, Free University of Berlin, Germany.
At Enzene, Mandar heads Discovery Operations, leading the team in strategies to develop discovery solutions and promoting efficient project execution.
Appalaraju Jaggupilli, Ph.D.
Senior Principal Scientist, Discovery Ops
Dr. Appalaraju Jaggupilli is a seasoned cell biologist with over 7 years of experience in protein biochemistry. He brings profound expertise in antibody and reagent protein production, drug discovery, and bioinformatics. Before joining Enzene Biosciences, he served as a Senior Research Investigator at Syngene International Ltd. and as a Postdoctoral Fellow at MD Anderson Cancer Center, where he successfully established mammalian expression systems and advanced drug discovery workflows.
He holds a Master’s degree in Biotechnology from the University of Salford, Manchester (UK), and a Ph.D. in Biochemistry from the University of Manitoba, Winnipeg (Canada). He also holds a Postgraduate Program in General Management from IIM Visakhapatnam, India.
At Enzene, Appalaraju leads the reagent protein production vertical, driving cross-functional collaborations within CEPA workflows and designing strategies for efficient protein expression and purification across diverse formats.
Rajeshwari B R, Ph.D.
Senior Scientist, Integrated Discovery
Dr. Rajeshwari B R specializes in cell and molecular biology. At Enzene, she works on developing integrated discovery platforms for recombinant protein expression, purification and functional characterization. She holds a Master’s degree in Biotechnology from M S Ramaiah Institute of Technology, Bangalore and a Ph.D. in Cell biology from IISER Pune.
Image Credit: © Rosianna - stock.adobe.com